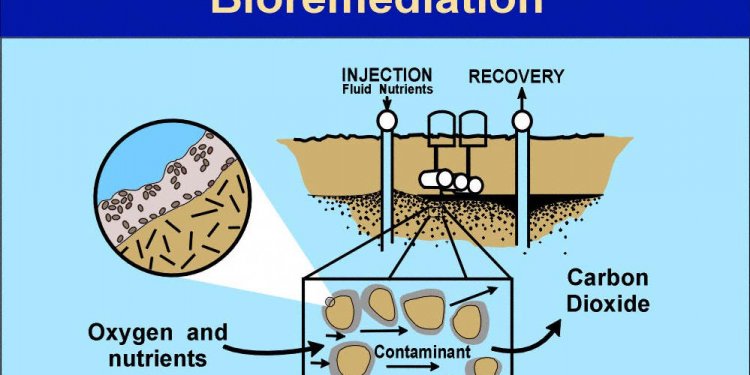
Microbial Bioremediation of oil spills
- U.S. Ecological Coverage Department, Cincinnati, Ohio 452682;
- Microbial Insights Inc., Rockford, Tennessee ; and
- Biological Sciences Division, Oak Ridge Nationwide Laboratory, Oak Ridge, Tennessee 378314
ABSTRACT
Three crude oil bioremediation practices had been used in a randomized block area test simulating a coastal oil spill. Four treatments (no oil control, oil alone, oil plus nutrients, and oil plus nutrients plus an indigenous inoculum) were applied. In situ microbial community structures were administered by phospholipid fatty acid (PLFA) analysis and 16S rDNA PCR-denaturing gradient serum electrophoresis (DGGE) to (i) identify the microbial community people in charge of the decontamination associated with website and (ii) establish a conclusion point the elimination of the hydrocarbon substrate. The outcomes of PLFA evaluation demonstrated a residential area change throughout plots from mainly eukaryotic biomass to gram-negative microbial biomass with time. PLFA pages from the oiled plots suggested increased gram-negative biomass and version to metabolic tension in comparison to unoiled controls. DGGE evaluation of untreated control plots revealed a straightforward, dynamic prominent populace construction through the experiment. This banding structure disappeared throughout oiled plots, showing the construction and variety of the prominent microbial community changed substantially. No constant distinctions had been detected between nutrient-amended and native inoculum-treated plots, but both differed from oil-only plots. Prominent rings had been excised for series evaluation and suggested that oil treatment encouraged the growth of gram-negative microorganisms inside the α-proteobacteria andFlexibacter-Cytophaga-Bacteroides phylum. α-Proteobacteria were never ever recognized in unoiled settings. PLFA analysis indicated that by week 14 the microbial community frameworks of this oiled plots were getting much like those associated with unoiled controls through the same time point, but DGGE analysis recommended that significant variations in the bacterial communities remained.
FOOTNOTES
- Received 1 March 1999.
- Corresponding author. Mailing address: U.S. ecological cover Agency, 26 W. Martin Luther King Dr., Cincinnati, OH 45268. Phone: (513) 569-7668. Fax: (513) 569-7105. E-mail: venosa.albert{at}epamail.epa.gov.